From foundation models like Evo 2 and Alphafold 3 to synthetic data generation and agentic AI workflows — Next‑gen AI tools are revolutionizing biomedical research. A fascinating question hung in the air at GTC 2025: How will these technologies reshape the role of human scientists? These aren’t just faster tools — they’re completely changing how we discover new medicines.

The AI-Powered Drug Discovery Revolution
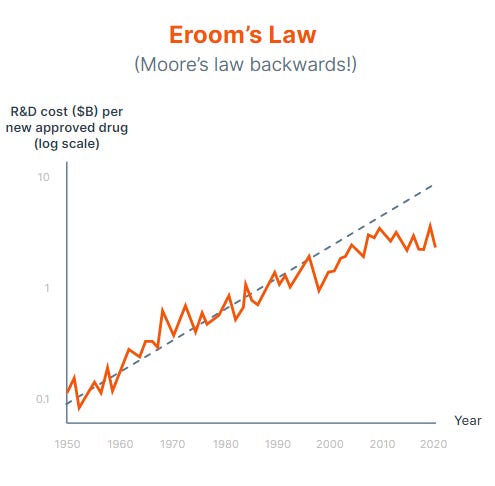
Drug discovery has traditionally been an arduous, expensive process, with the average therapeutic taking 10–15 years and billions of dollars to reach market. The integration of foundation models and specialized AI infrastructure is reshaping this landscape, enabling pharmaceutical companies to screen billions of compounds in silico before moving to laboratory testing.
As emphasized at GTC, NVIDIA’s BioNeMo platform serves as a cornerstone for these advances, offering optimized performance for biological and chemical model training and inference. BioNeMo provides a comprehensive ecosystem for drug discovery teams, with performance metrics significantly outpacing standard implementations:
AlphaFold2 runs 5x faster as a BioNeMo NVIDIA Inference Microservices (NIM) than standard implementations
DiffDock 2.0 achieves 6.2x speed-up on molecular orientation prediction
Molecular generation models process tokens significantly faster compared to baselines
Expert Perspectives on Foundation Models in Biology
Foundation models in biology represent a paradigm shift, enabling researchers to work across different layers of biological meaning and length scales of abstraction.

Molly Gibson, Co-Founder of Lila Sciences — the Flagship Pioneering startup chasing scientific super-intelligence which recently came out of stealth — highlighted that while these models offer powerful capabilities, the results depend heavily on who’s using them: “Different scientists using the same foundation model can produce dramatically different results,” she explained, emphasizing that junior researchers often struggle to properly evaluate model outputs, making them less productive than their senior counterparts, who already possess the knowledge needed to separate the wheat from the chaff. The critical question won’t be whether AI will replace scientists, but rather:
“Who’s going to be the best users of these systems and how does that change over time?”
Patrick Hsu, fresh off the announcement of Evo 2 — a 40-billion-parameter open-source AI model trained on 9.3 trillion nucleotides from over 128,000 genomes — stressed the importance of models that bridge across “different layers of biological meaning and length scales of abstraction,” while maintaining explainable reasoning chains.
Evo 2 stands out for its ability to analyze and generate entire genomic sequences across all domains of life, predict the effects of genetic mutations, and integrate DNA, RNA, and protein data, enabling multi-scale biological insights that distinguish foundation models from their more specialized predecessors. Evo 2 has set a new benchmark in biology by achieving state-of-the-art accuracy in predicting both coding and noncoding mutation effects, and by enabling the design of complex genomes and targeted genetic therapies.
Josh traced the evolution of the field from specialized models to fine-tuned approaches and now to powerful foundation models used directly. He emphasized that for practical drug development, models must “grasp whether the drug would be manufacturable,” not just predict its efficacy — a principle that guides Chai Discovery as they integrate manufacturability assessments into their AI-driven pipeline.
Nick characterized foundation models in biology as “distributional learners,” noting the challenges in developing effective prompting strategies for biological applications — a focus at EvolutionaryScale, where he contributes to advancing models like ESM3 that leverage prompting and chain-of-thought reasoning to engineer novel proteins and biological functions.
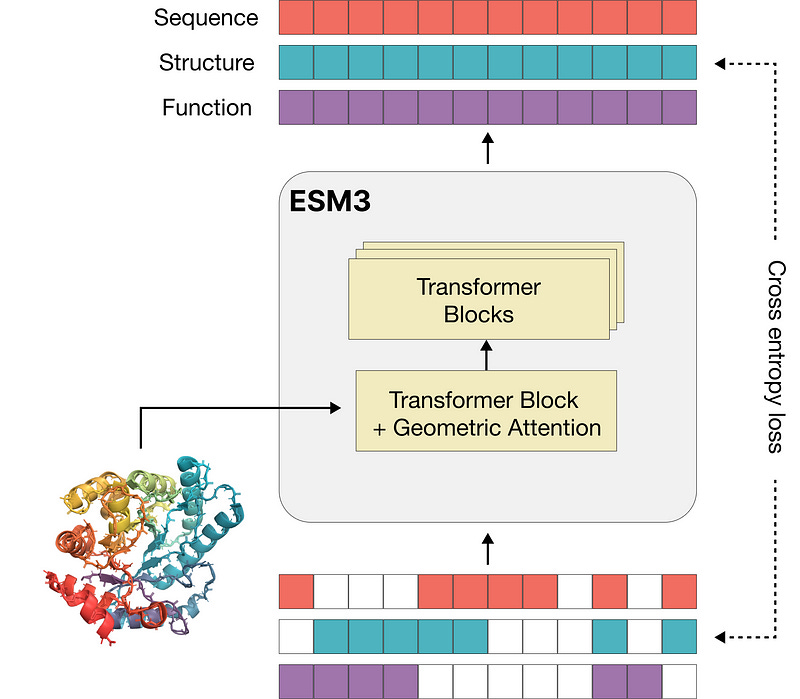
Several panelists, including Patrick, advocated for open-source approaches to build ecosystem trust, pointing to Evo 2 as an open source alternative to closed vendor systems. A persistent challenge remains what Molly identified as data imbalance, with “all the data in early discovery stages” while “data in later stages are harder to get access to.”
Protein Structure Prediction with AlphaFold 3
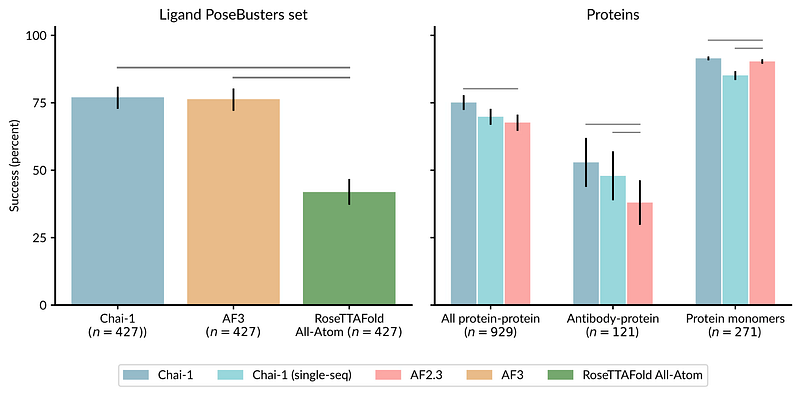
AlphaFold 3, developed by Isomorphic Labs (spun out of Google DeepMind), predicts protein-DNA and molecular interactions with impressive accuracy. While built on AlphaFold 2’s core principles, AlphaFold 3’s Pairformer + diffusion architecture marks a conceptual shift toward unified, generative modelling of all life’s molecules, delivering both higher accuracy and richer uncertainty estimates for complex assemblies.
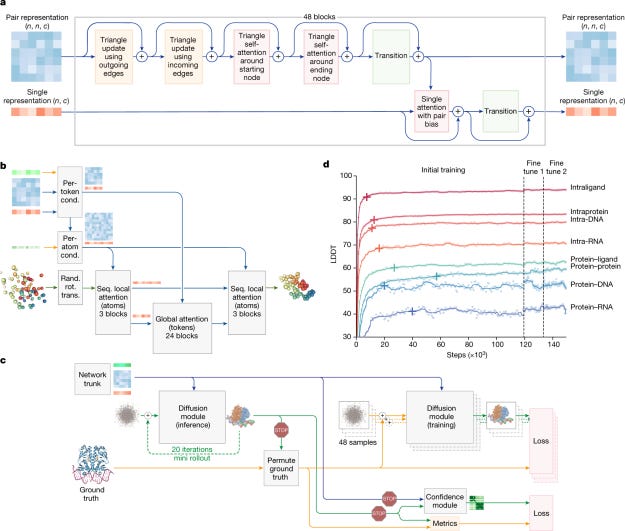
AlphaFold 3 introduced a diffusion-based architecture that predicts structures of diverse biomolecular complexes — including proteins, nucleic acids, ligands, and ions — with significantly higher accuracy than prior models. It outperformed previous state-of-the-art tools in protein–ligand docking, nucleic acid interactions, and antibody–antigen prediction, demonstrating that a single deep learning model can achieve high-accuracy structural modeling across biomolecular types.

From In-Silico towards In-Vivo:
Sergei highlighted the full cycle from AI in-silico agents to experimental validation:
AI agents query molecular databases
AI analogues predict interactions and properties
Laboratory observations validate predictions
Results inform the next generation of queries
The isomorphic mapping between the world of biology and the world of AI creates powerful synergies. The goal, as described by Jaderberg, is to eventually conduct “entire drug design campaigns in one continuous round” rather than the fragmented approach currently used.
NVIDIA BioNeMo Adoption
In collab with NVIDIA, several companies announced generative AI tools for molecular design at GTC:
Sapio Sciences announced a no-code informatics platform integrating NVIDIA BioNeMo.
“Integrating BioNeMo into Sapio’s AI-driven research platform gives scientists access to advanced generative AI models for drug discovery. With AlphaFold2, MoIMIM, and DiffDock NIMs, researchers can predict, optimize, and validate drug candidates with greater speed and accuracy. This work underscores AI’s growing role in transforming pharmaceutical research and accelerating the path to breakthrough treatments.” — Anthony Costa, Director, Digital Biology at NVIDIA
Cadence announced their cloud-native Orion molecular design platform will also be supercharged with BioNeMo’s capabilities.
These foundation models enable scientists to systematically explore vast chemical spaces, predict protein-ligand interactions, and identify promising drug candidates with targeted therapeutic properties.
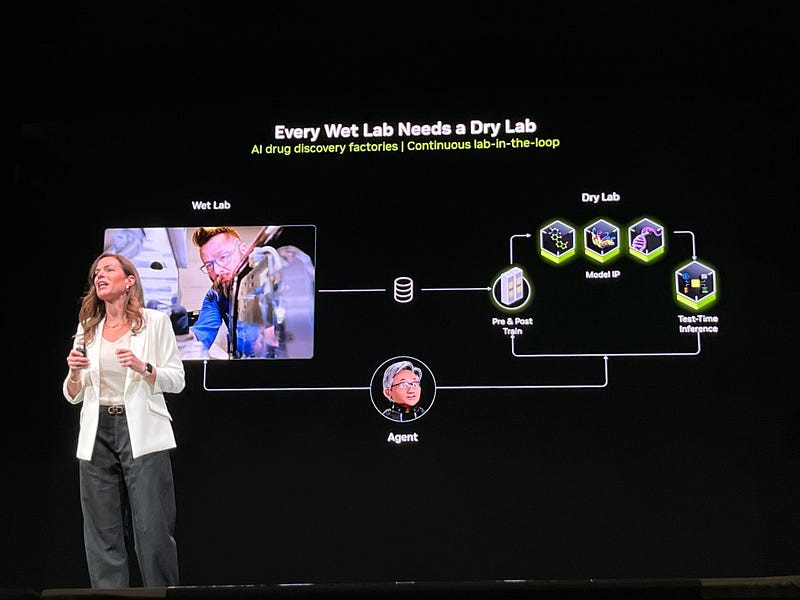
The Continuous AI Factory Approach for Drug Discovery
Kimberly Powell introduced the concept of AI drug discovery factories where every wet lab needs a complementary dry lab. This approach creates a continuous feedback loop between computational prediction and experimental validation:
AI models generate candidate molecules
Laboratory testing provides validation data
AI Agent Jensen-Huang-in-your-computer processes data
Results feed back to improve AI models
Process repeats with enhanced accuracy
This iterative approach has already demonstrated impressive results. Companies like Daiichi-Sankyo have screened 6 billion molecules, while Schrodinger has evaluated 8.2 billion compounds using these accelerated methods.
NVIDIA’s Technological Stack for Drug Discovery
The conference highlighted NVIDIA’s comprehensive technology stack supporting drug discovery:
Hardware Infrastructure
DGX systems for AI training
Grace Blackwell architecture for high-performance computing
Specialized memory bandwidth (576 TB/second in NVLink72 systems) essential for complex biological modeling
Software Ecosystem
NVIDIA BioNeMo platform optimized for biological applications. Train and deploy LLMs, or access them through an inference-optimized api key through BioNeMo NIMs
MONAI (Medical Open Network for AI) PyTorch-based open-source framework with 4.5M downloads, and 35+ models for medical imaging
Specialized NIMs for genomics and structural biology
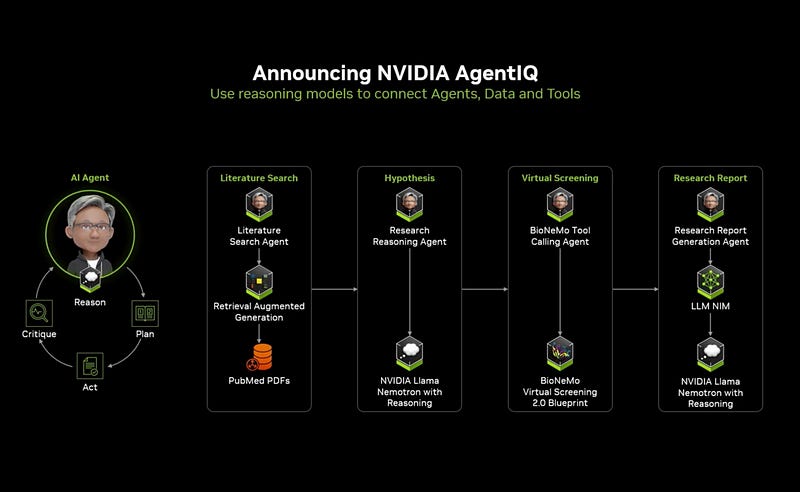
Agent frameworks enabling autonomous research exploration
The Future: Agentic Drug Discovery
The most forward-looking discussions centered on fully autonomous AI agents for drug discovery. These systems, powered by reasoning capabilities of large language models like NVIDIA’s LLaMa NemoTron, can:
Design small molecules and antibodies with minimal human guidance
Optimize compounds for specific protein interactions
Generate and test hypotheses independently
Scale experimentation from one chemist to “thousands of agents”
Notable companies exploring this frontier include Humanate for patient intake, Accenture for clinical trial design, and Quantiphi for clinical development.
So what?
The integration of AI into drug discovery represents more than incremental improvement — it’s a fundamental reimagining of how we develop therapeutics. By leveraging specialized hardware, optimized software frameworks, and increasingly autonomous AI systems, researchers can explore biological systems with unprecedented speed and precision. The future of drug discovery is computational and collaborative — driven by both human expertise and machine intelligence.
Welcome to Sprintome
Drug discovery is getting an upgrade.
At Sprintome, we focus on the quiet revolutions in drug discovery — measurable, verifiable steps that compound over time. No shortcuts, just steady acceleration.
Join the sprint.